Introduction
Sampling without replacement implies that elements of a population can be chosen only once in one sample. There are different techniques of sampling without replacement, including:
- simple random sampling
- random sampling with unequal probabilities
- stratified sampling
- balanced sampling (a special case of stratified sampling)
- permutation sampling
- k-fold sampling
- leave-p-out sampling
In this article, we show how to draw repeated samples without
replacement from a population by using the functions
implemented in resampling.
For further information on the sampling techniques, see Sampling without replacement.
Setup
Loading
Firstly, we need to load resampling and other needed R
packages:
#resampling
library(resampling)
#Packages for visualisation
require(ComplexHeatmap, quietly = TRUE)
require(grid, quietly = TRUE)
require(RColorBrewer, quietly = TRUE)Seed
Then, we set a seed for the random number generation (RNG). In fact,
different R sessions have different seeds created from current time and
process ID by default, and consequently different simulation results. By
fixing a seed we ensure we will be able to reproduce the results of this
vignette. We can specify a seed by calling ?set.seed.
#Set a seed for RNG
set.seed(
#A seed
seed = 5381L, #a randomly chosen integer value
#The kind of RNG to use
kind = "Mersenne-Twister", #we make explicit the current R default value
#The kind of Normal generation
normal.kind = "Inversion" #we make explicit the current R default value
)Resampling Without Replacement
The available methods for taking repeated samples without replacement
can be listed through the ?listAvailableSamplingMethods
function call, setting the input argument to 'rswor'.
?listAvailableSamplingMethods returns a table with two
columns:
-
id: the id of the sampling method, to be used in the function calls -
name: the name of the sampling method
#list sampling methods
sampling.methods = listAvailableSamplingMethods(x = 'rswor')
#print in table
knitr::kable(x = sampling.methods)| id | name |
|---|---|
| rswor | random sampling without replacement |
| srswor | simple random sampling without replacement |
| stratified_rswor | stratified random sampling without replacement |
| balanced_rswor | balanced random sampling without replacement |
| permutation | permutation sampling |
| kfolds | random k-fold sampling |
| stratified_kfolds | stratified k-fold sampling |
| balanced_kfolds | balanced k-fold sampling |
| leave_p_out | leave-p-out sampling |
| leave_one_out | leave-one-out sampling |
The name of the resampling functions can be retrieved by calling
?listResamplingFunctionNames.
#list resampling function names
resampling.function.names = listResamplingFunctionNames(x = 'rswor')
#print in table
knitr::kable(x = resampling.function.names)| id | name |
|---|---|
| rswor | repeatedSampleWithoutReplacement |
| srswor | repeatedSimpleRandomSampleWithoutReplacement |
| stratified_rswor | repeatedStratifiedSampleWithoutReplacement |
| balanced_rswor | repeatedBalancedSampleWithoutReplacement |
| permutation | repeatedPermutationSample |
| kfolds | repeatedRandomKm1Folds |
| stratified_kfolds | repeatedStratifiedKm1Folds |
| balanced_kfolds | repeatedBalancedKm1Folds |
| leave_p_out | repeatedLeavePOutSample |
| leave_one_out | repeatedLeaveOneOutSample |
Each function is documented. To learn more about a specific method it
is possible to use the ? operator. For example, let’s check
the function
?repeatedSimpleRandomSampleWithoutReplacement.
#See documentation
?repeatedSimpleRandomSampleWithoutReplacementFrom the documentation, we can see that the function accepts 3 arguments in input:
-
k: the number of repeated samples to generate -
N: the population size -
n: the sample size
Simple Random Sampling
In resampling via simple random sampling (SRS), simple random samples without replacement are repeatedly taken from the population.
The function implementing this sampling scheme is
?repeatedSimpleRandomSampleWithoutReplacement, which
accepts 3 arguments:
-
k: the number of repeated samples to generate -
N: the population size -
n: the sample size
#Simple random sampling without replacement
repeatedSimpleRandomSampleWithoutReplacement(
k = 2,
N = 10,
n = 8
)
#> [[1]]
#> [1] 1 9 7 2 4 3 6 10
#>
#> [[2]]
#> [1] 9 8 1 7 4 5 6 3Instead of using
?repeatedSimpleRandomSampleWithoutReplacement, we can take
repeated samples by using the ?resample function.
#Simple random sampling without replacement
obj = resample(
x = 10,
n = 8,
k = 2,
method = 'srswor'
)
#Print
print(obj)
#>
#> 2 samples taken from a population of 10 elements by using simple random
#> sampling without replacement.
#>
#> sampleNumber sample sampleSize holdoutSize
#> 1 1 2, 3, 9, 5, ... 8 2
#> 2 2 2, 10, 8, , ... 8 2
#Samples
getSamples(obj)
#> [[1]]
#> [1] 2 3 9 5 7 4 6 1
#>
#> [[2]]
#> [1] 2 10 8 1 6 4 3 7
#Holdout Data
getHoldOutSamples(obj)
#> [[1]]
#> [1] 8 10
#>
#> [[2]]
#> [1] 5 9
#Plot
plot(x = obj)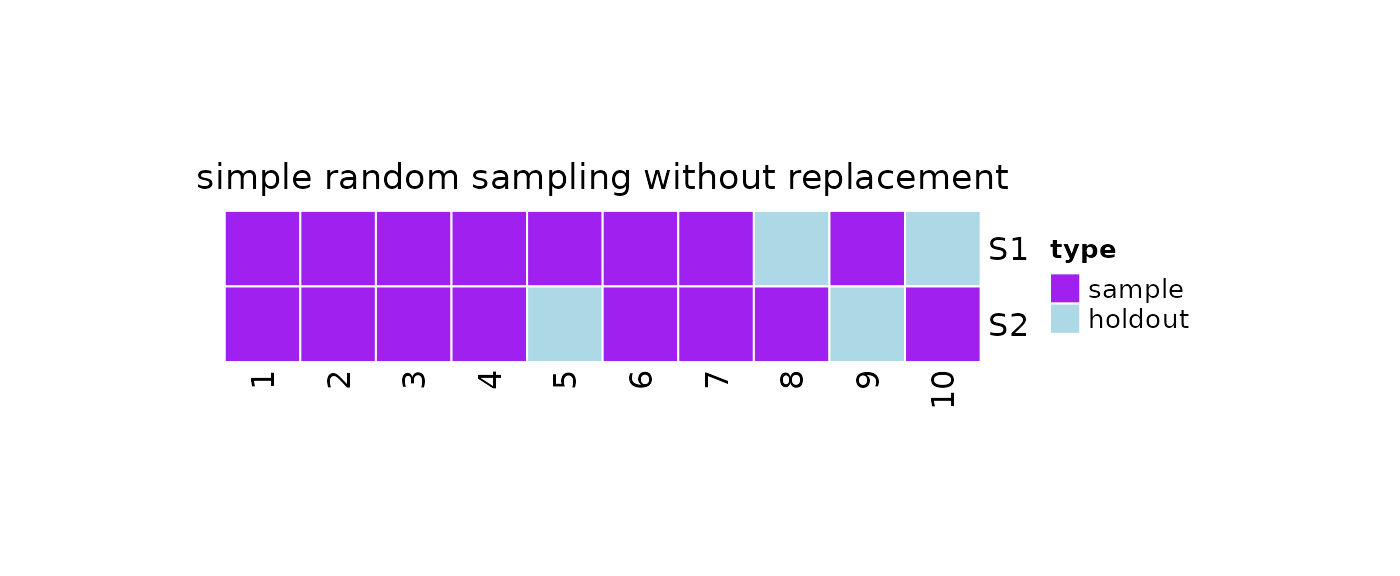
Random Sampling With Unequal Probability
We can use ?repeatedSampleWithoutReplacement to draw
repeated samples without replacement with unequal
probability. From the documentation, we can see that the
function accepts 4 arguments in input:
-
k: the number of repeated samples to generate -
N: the population size -
n: the sample size -
prob: an optional vector of probabilities for obtaining the population elements
For example, let’s assume our population of interest has 10 elements, and that the first 3 elements have an higher chance of being selected.
#Random sampling without replacement
repeatedSampleWithoutReplacement(
k = 2,
N = 10,
n = 5,
prob = c(rep(3,3), rep(1,7))
)
#> [[1]]
#> [1] 3 7 10 2 1
#>
#> [[2]]
#> [1] 7 2 1 9 3We can take repeated samples by using the ?resample
function and setting method = 'rswor'.
#Random sampling without replacement
obj = resample(
x = 10,
n = 5,
k = 2,
method = 'rswor',
prob = c(rep(3,3),rep(1,7))
)
#Print
print(obj)
#>
#> 2 samples taken from a population of 10 elements by using random
#> sampling without replacement.
#>
#> sampleNumber sample sampleSize holdoutSize
#> 1 1 7, 3, 6, 2, ... 5 5
#> 2 2 6, 1, 2, 3, ... 5 5
#Samples
getSamples(obj)
#> [[1]]
#> [1] 7 3 6 2 1
#>
#> [[2]]
#> [1] 6 1 2 3 8
#Holdout Data
getHoldOutSamples(obj)
#> [[1]]
#> [1] 4 5 8 9 10
#>
#> [[2]]
#> [1] 4 5 7 9 10
#Plot
plot(x = obj)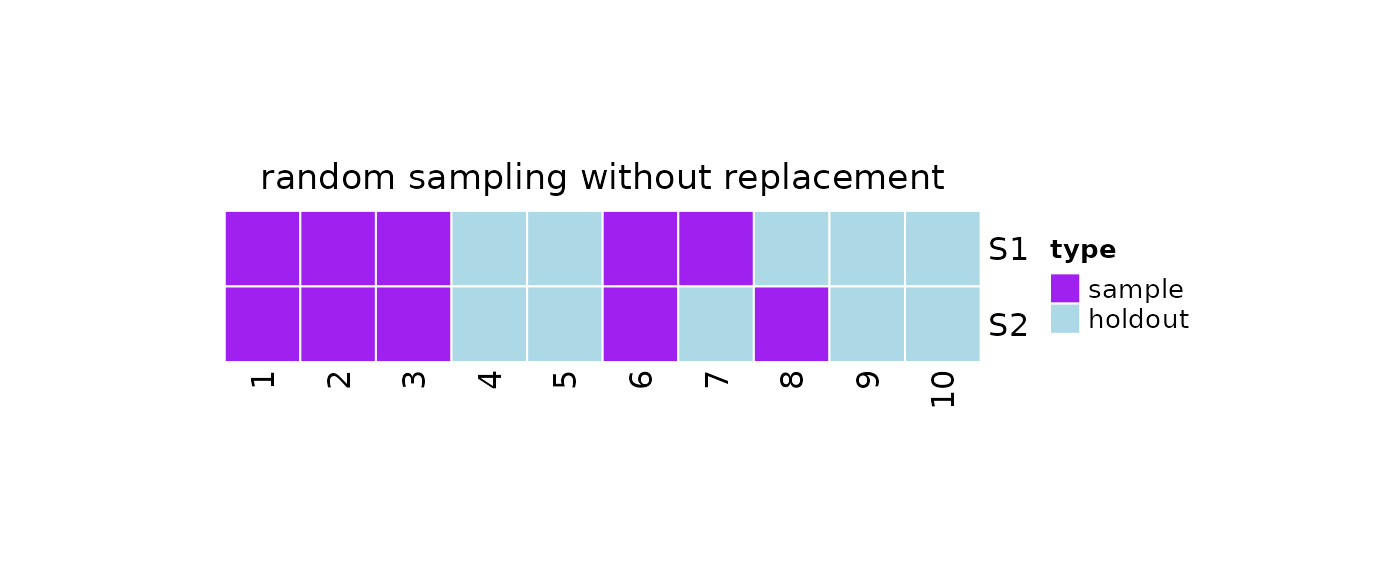
Stratified Random Sampling
Repeated stratified samples without replacement can
be taken by using
?repeatedStratifiedSampleWithoutReplacement which accept a
strata argument in input.
#Define strata
strata = c(rep("a", 3),rep("b", 6))
#Stratified sampling without replacement
repeatedStratifiedSampleWithoutReplacement(
k = 2,
strata = strata,
n = 6
)
#> [[1]]
#> [1] 2 1 4 9 6 5
#>
#> [[2]]
#> [1] 3 5 4 6 2 9We can take repeated samples by using the ?resample
function and setting method = 'stratified_rswor'.
#Stratified sampling without replacement
obj = resample(
x = strata,
n = 6,
k = 2,
method = 'stratified_rswor'
)
#Print
print(obj)
#>
#> 2 samples taken from a population of 9 elements by using stratified
#> random sampling without replacement.
#>
#> sampleNumber sample sampleSize holdoutSize
#> 1 1 1, 7, 5, 8, ... 6 3
#> 2 2 4, 8, 9, 2, ... 6 3
#Samples
getSamples(obj)
#> [[1]]
#> [1] 1 7 5 8 4 2
#>
#> [[2]]
#> [1] 4 8 9 2 5 1
#Holdout Data
getHoldOutSamples(obj)
#> [[1]]
#> [1] 3 6 9
#>
#> [[2]]
#> [1] 3 6 7
#Plot
plot(x = obj, strata = strata)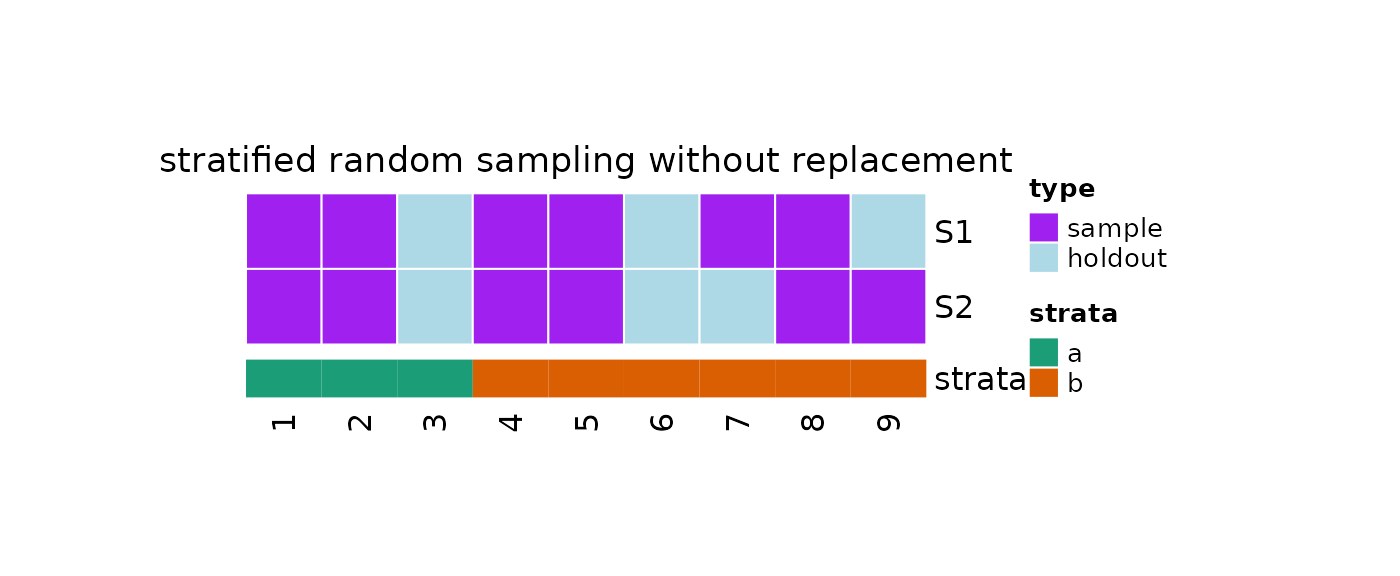
Balanced Random Sampling
Balanced sampling is a special case of stratified sampling used to ensure that subgroups of the population are equally represented in the taken sample.
Repeated balanced samples without replacement can be taken by using
?repeatedBalancedSampleWithoutReplacement:
#Define strata
strata = c(rep("a", 3),rep("b", 6))
#Check ratio
table(strata)/length(strata)
#> strata
#> a b
#> 0.3333333 0.6666667
#Balanced sampling without replacement
s = repeatedBalancedSampleWithoutReplacement(
k = 2,
strata = strata,
n = 6
)
#Check ratio in the samples
table(strata[s[[1]]])/length(strata[s[[1]]])
#>
#> a b
#> 0.5 0.5
table(strata[s[[2]]])/length(strata[s[[2]]])
#>
#> a b
#> 0.5 0.5We can take repeated samples by using the ?resample
function and setting method = 'balanced_rswor'.
#Balanced sampling without replacement
obj = resample(
x = strata,
n = 6,
k = 2,
method = 'balanced_rswor'
)
#Print
print(obj)
#>
#> 2 samples taken from a population of 9 elements by using balanced
#> random sampling without replacement.
#>
#> sampleNumber sample sampleSize holdoutSize
#> 1 1 8, 9, 3, 7, ... 6 3
#> 2 2 1, 5, 2, 3, ... 6 3
#Samples
getSamples(obj)
#> [[1]]
#> [1] 8 9 3 7 1 2
#>
#> [[2]]
#> [1] 1 5 2 3 7 4
#Holdout Data
getHoldOutSamples(obj)
#> [[1]]
#> [1] 4 5 6
#>
#> [[2]]
#> [1] 6 8 9
#Plot
plot(x = obj, strata = strata)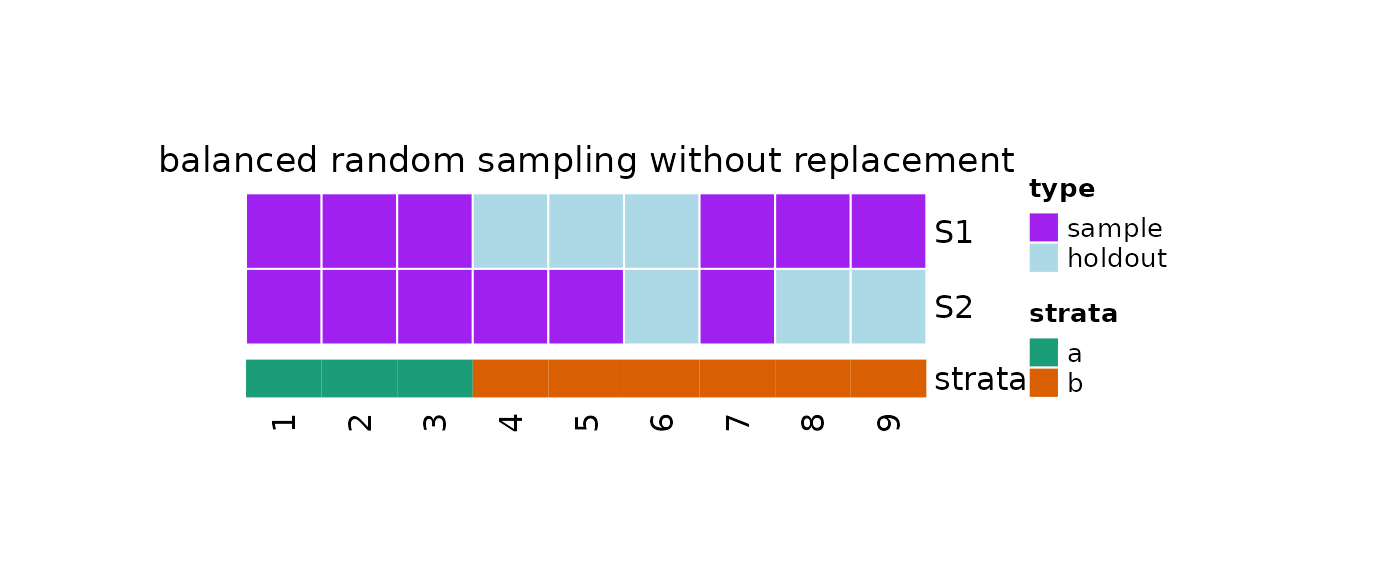
Permutation Sampling
We can take repeated permutation samples by using
?repeatedPermutationSample:
#Permutation sampling
repeatedPermutationSample(
k = 2,
N = 10
)
#> [[1]]
#> [1] 3 6 8 10 2 1 5 9 7 4
#>
#> [[2]]
#> [1] 9 7 2 1 5 8 4 6 3 10We can also use the ?resample function by setting
method = 'permutation'.
#Permutation sampling
obj = resample(
x = 10,
k = 2,
method = 'permutation'
)
#Print
print(obj)
#>
#> 2 samples taken from a population of 10 elements by using permutation
#> sampling.
#>
#> sampleNumber sample sampleSize holdoutSize
#> 1 1 10, 3, 5, , ... 10 0
#> 2 2 3, 9, 5, 4, ... 10 0
#Samples
getSamples(obj)
#> [[1]]
#> [1] 10 3 5 8 7 6 4 2 9 1
#>
#> [[2]]
#> [1] 3 9 5 4 8 10 1 6 2 7
#Holdout Data
getHoldOutSamples(obj)
#> [[1]]
#> integer(0)
#>
#> [[2]]
#> integer(0)
#Plot
plot(x = obj)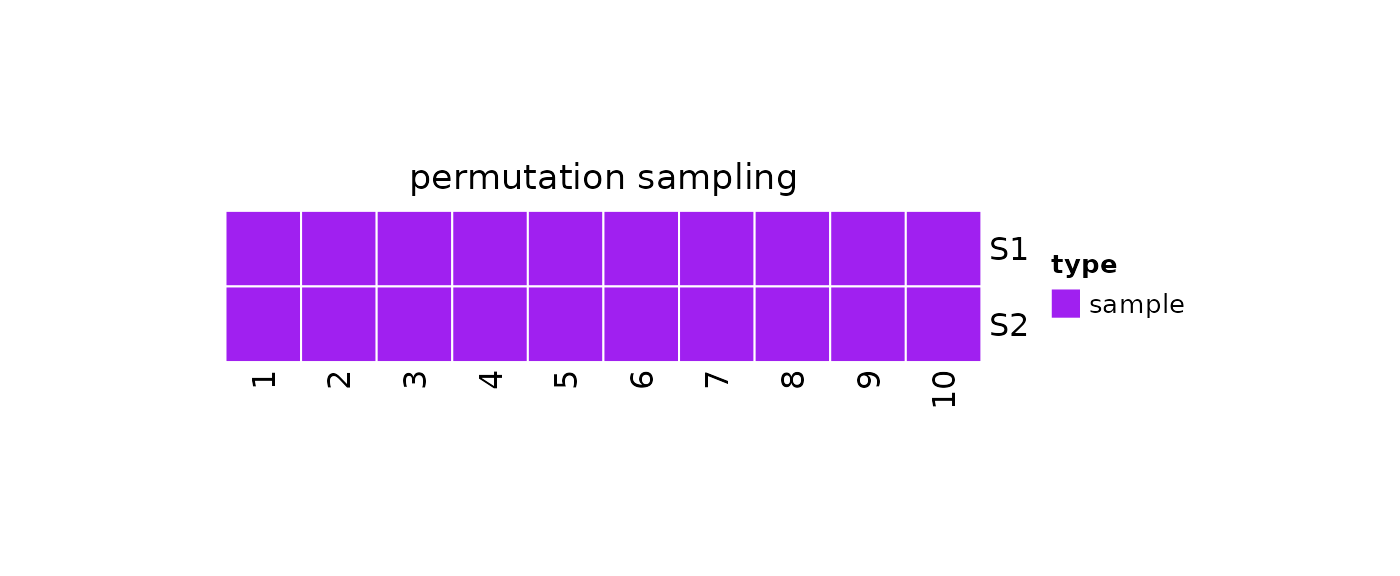
K-fold Sampling
The idea behind k-fold resampling is taken from k-fold cross-validation: k samples are taken from the population, so that the i-th sample is generated by removing the i-th fold from the population and merging the remaining k - 1 folds together.
We can take k samples via k-fold sampling by calling
?repeatedRandomKm1Folds. The function accepts two arguments
in input:
-
k: the number of folds -
N: the population size
?repeatedRandomKm1Folds returns a list of length
k where each element is a sample obtained by merging
k-1 folds together.
#K-1 folds sampling
repeatedRandomKm1Folds(
N = 10,
k = 3
)
#> [[1]]
#> [1] 1 2 3 4 7 8
#>
#> [[2]]
#> [1] 3 4 5 6 8 9 10
#>
#> [[3]]
#> [1] 1 2 5 6 7 9 10We can take repeated samples by using the ?resample
function and setting method = 'kfolds'.
#K-folds sampling
obj = resample(
x = 10,
k = 3,
method = "kfolds"
)
#Print
print(obj)
#>
#> 3 samples taken from a population of 10 elements by using random k-fold
#> sampling.
#>
#> sampleNumber sample sampleSize holdoutSize
#> 1 1 1, 3, 4, 7, ... 6 4
#> 2 2 2, 3, 4, 5, ... 7 3
#> 3 3 1, 2, 5, 6, ... 7 3
#Samples
getSamples(obj)
#> [[1]]
#> [1] 1 3 4 7 8 10
#>
#> [[2]]
#> [1] 2 3 4 5 6 8 9
#>
#> [[3]]
#> [1] 1 2 5 6 7 9 10
#Holdout Data
getHoldOutSamples(obj)
#> [[1]]
#> [1] 2 5 6 9
#>
#> [[2]]
#> [1] 1 7 10
#>
#> [[3]]
#> [1] 3 4 8
#Plot
plot(obj)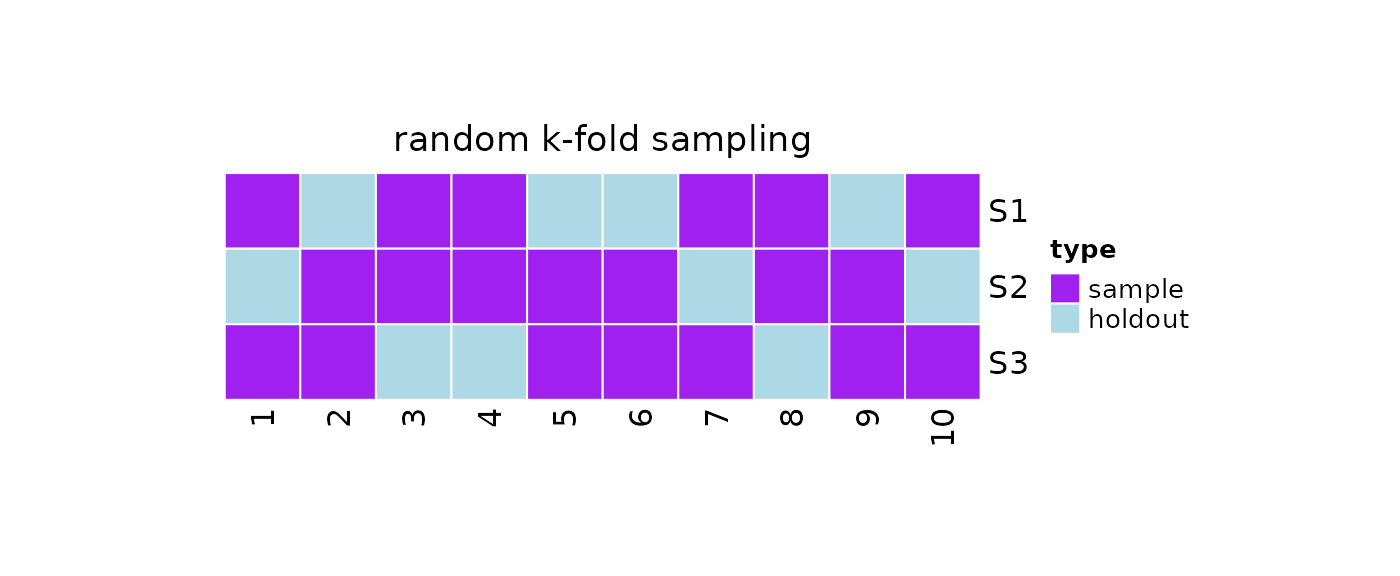
Stratified K-fold Sampling
We can take repeated stratified samples via stratified k-fold
sampling by calling
?repeatedStratifiedKm1Folds.
?repeatedStratifiedKm1Folds assigns the population to
k stratified folds and returns k samples made of
elements from k-1 folds:
#Define strata
strata = c(rep("a", 3),rep("b", 6))
#Check ratio
table(strata)/length(strata)
#> strata
#> a b
#> 0.3333333 0.6666667
#Assign data to 3 folds
i = repeatedStratifiedKm1Folds(
strata = strata,
k = 3
)
#Check folds
i
#> [[1]]
#> [1] 1 2 5 7 8 9
#>
#> [[2]]
#> [1] 2 3 4 5 6 8
#>
#> [[3]]
#> [1] 1 3 4 6 7 9
#Check ratio in the folds
table(strata[i[[1]]])/length(strata[i[[1]]])
#>
#> a b
#> 0.3333333 0.6666667
table(strata[i[[2]]])/length(strata[i[[2]]])
#>
#> a b
#> 0.3333333 0.6666667
table(strata[i[[3]]])/length(strata[i[[3]]])
#>
#> a b
#> 0.3333333 0.6666667We can take repeated samples by using the ?resample
function and setting method = 'stratified_kfolds'.
#Define strata
strata = c(rep("a", 3),rep("b", 6))
#Stratified k-folds sampling
obj = resample(
x = strata,
k = 3,
method = "stratified_kfolds"
)
#Print
print(obj)
#>
#> 3 samples taken from a population of 9 elements by using stratified
#> k-fold sampling.
#>
#> sampleNumber sample sampleSize holdoutSize
#> 1 1 1, 3, 4, 5, ... 6 3
#> 2 2 1, 2, 5, 6, ... 6 3
#> 3 3 2, 3, 4, 7, ... 6 3
#Samples
getSamples(obj)
#> [[1]]
#> [1] 1 3 4 5 6 7
#>
#> [[2]]
#> [1] 1 2 5 6 8 9
#>
#> [[3]]
#> [1] 2 3 4 7 8 9
#Holdout Data
getHoldOutSamples(obj)
#> [[1]]
#> [1] 2 8 9
#>
#> [[2]]
#> [1] 3 4 7
#>
#> [[3]]
#> [1] 1 5 6
#Plot
plot(x = obj, strata = strata)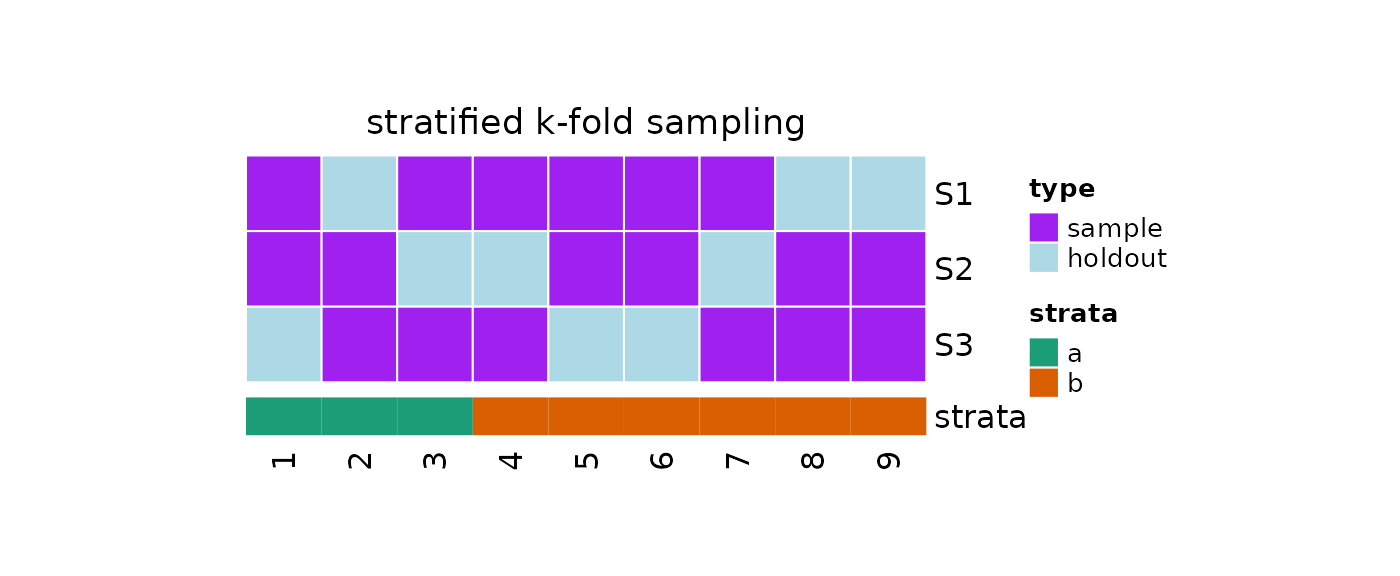
Balanced K-fold Sampling
We can take repeated balanced samples via balanced k-fold
sampling by calling ?repeatedBalancedKm1Folds,
which returns k samples made of elements from k-1
folds.
Internally, it uses ?balancedKFolds to assign the
population to k balanced folds:
- If the population is balanced, each element in the population is assigned to one of the k folds so that the percentage of each stratum is preserved in each fold
- If the population is unbalanced and
undersample = TRUE, the so-called random undersampling is adopted, i.e. the proportion of the strata in the population is adjusted by removing elements from the majority groups, so that each stratum is balanced
#Define balanced strata
strata = c(rep(1,6),rep(2,6))
#Check ratio
table(strata)/length(strata)
#> strata
#> 1 2
#> 0.5 0.5
#Assign data to 3 folds and take a sample
i = repeatedBalancedKm1Folds(
strata = strata,
k = 3
)
#Check sample
i
#> [[1]]
#> [1] 1 2 4 5 7 8 9 10
#>
#> [[2]]
#> [1] 2 3 4 6 8 9 11 12
#>
#> [[3]]
#> [1] 1 3 5 6 7 10 11 12
#Check ratio in the sample
table(strata[i[[1]]])/length(strata[i[[1]]])
#>
#> 1 2
#> 0.5 0.5
table(strata[i[[2]]])/length(strata[i[[2]]])
#>
#> 1 2
#> 0.5 0.5
table(strata[i[[3]]])/length(strata[i[[3]]])
#>
#> 1 2
#> 0.5 0.5
#Define unbalanced strata
strata = c(rep(1,6),rep(2,12))
#Check ratio
table(strata)/length(strata)
#> strata
#> 1 2
#> 0.3333333 0.6666667
#Assign data to 3 folds and take a sample
i = repeatedBalancedKm1Folds(
strata = strata,
k = 3,
undersample = T
)
#Check folds
i
#> [[1]]
#> [1] 3 4 5 6 8 13 17 18
#>
#> [[2]]
#> [1] 1 2 4 6 8 12 14 18
#>
#> [[3]]
#> [1] 1 2 3 5 12 13 14 17
#>
#> attr(,"removed.data")
#> [1] 7 9 10 11 15 16
#Check ratio in the folds
table(strata[i[[1]]])/length(strata[i[[1]]])
#>
#> 1 2
#> 0.5 0.5
table(strata[i[[2]]])/length(strata[i[[2]]])
#>
#> 1 2
#> 0.5 0.5
table(strata[i[[3]]])/length(strata[i[[3]]])
#>
#> 1 2
#> 0.5 0.5We can take repeated samples by using the ?resample
function.
#Define strata
strata = c(rep("a", 6),rep("b", 6))
#Balanced k-folds sampling (balanced population)
obj = resample(
x = strata,
k = 3,
method = "balanced_kfolds"
)
#Print
print(obj)
#>
#> 3 samples taken from a population of 12 elements by using balanced
#> k-fold sampling.
#>
#> sampleNumber sample sampleSize holdoutSize
#> 1 1 1, 3, 4, 6, ... 8 4
#> 2 2 1, 2, 3, 5, ... 8 4
#> 3 3 2, 4, 5, 6, ... 8 4
#Samples
getSamples(obj)
#> [[1]]
#> [1] 1 3 4 6 7 9 10 12
#>
#> [[2]]
#> [1] 1 2 3 5 7 8 11 12
#>
#> [[3]]
#> [1] 2 4 5 6 8 9 10 11
#Holdout Data
getHoldOutSamples(obj)
#> [[1]]
#> [1] 2 5 8 11
#>
#> [[2]]
#> [1] 4 6 9 10
#>
#> [[3]]
#> [1] 1 3 7 12
#Plot
plot(x = obj, strata = strata)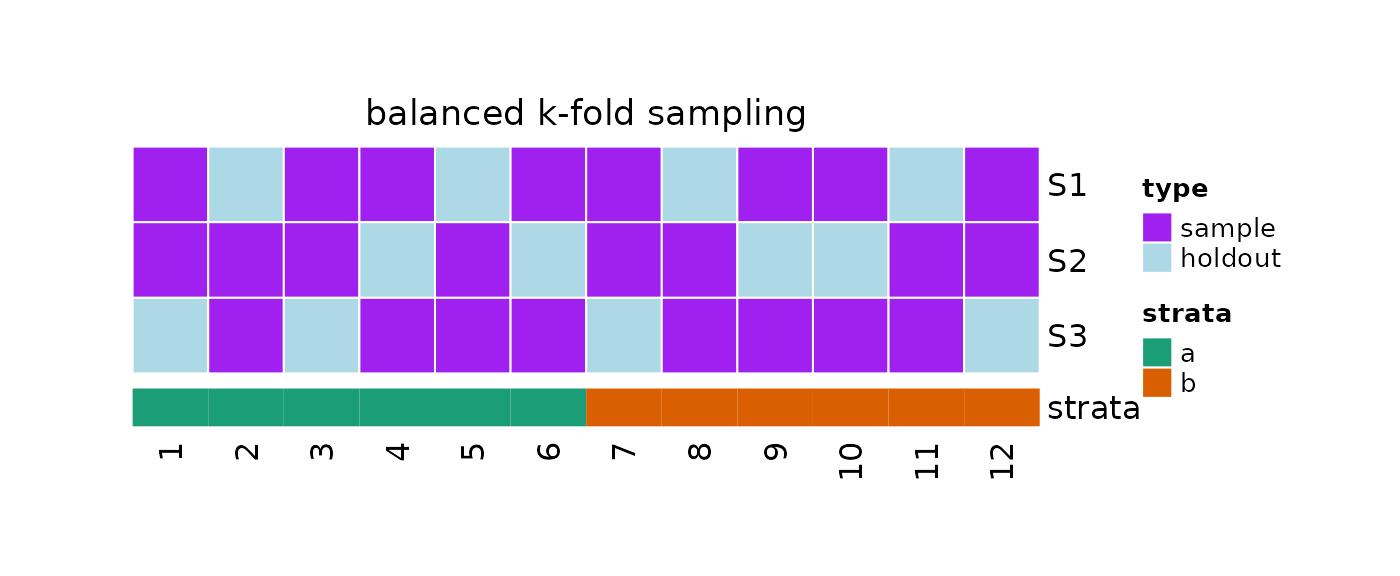
In case of unbalanced population, we can use the
undersample argument.
#Define strata
strata = c(rep("a",6),rep("b",8))
#Balanced k-folds sampling (unbalanced population)
obj = resample(
x = strata,
k = 3,
method = "balanced_kfolds",
undersample = T
)
#Print
print(obj)
#>
#> 3 samples taken from a population of 14 elements by using balanced
#> k-fold sampling.
#>
#> sampleNumber sample sampleSize holdoutSize
#> 1 1 1, 2, 3, 4, ... 8 4
#> 2 2 3, 4, 5, 6, ... 8 4
#> 3 3 1, 2, 5, 6, ... 8 4
#Samples
getSamples(obj)
#> [[1]]
#> [1] 1 2 3 4 7 10 12 13
#>
#> [[2]]
#> [1] 3 4 5 6 7 8 10 14
#>
#> [[3]]
#> [1] 1 2 5 6 8 12 13 14
#Holdout Data
getHoldOutSamples(obj)
#> [[1]]
#> [1] 5 6 8 14
#>
#> [[2]]
#> [1] 1 2 12 13
#>
#> [[3]]
#> [1] 3 4 7 10
#Plot
plot(x = obj, strata = strata)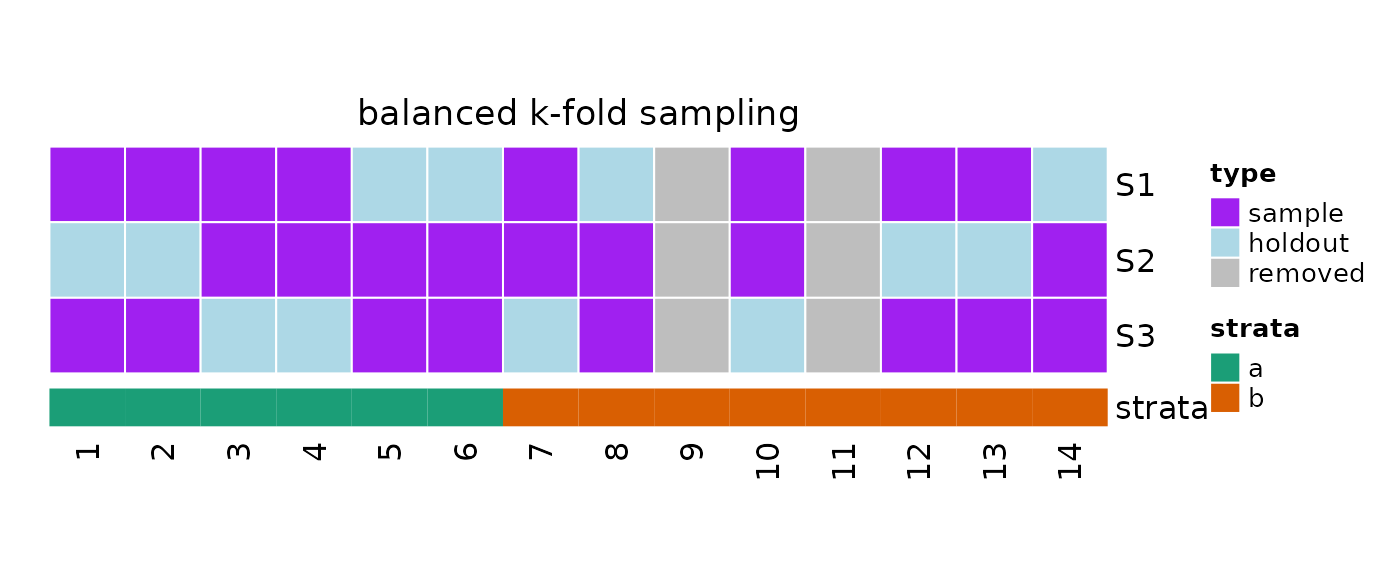
Leave-p-out Sampling
Leave-p-out resampling is an exhaustive resampling
technique, in which samples of size p are repeatedly taken
from the population until all the possible combinations of
p elements are considered. These samples are then used as
holdout data.
?repeatedLeavePOut returns a list of length \(\binom{N}{p}\) where each element is a
sample obtained by removing the holdout data:
#Repeatedly sample leaving out p elements each time
repeatedLeavePOut(N = 5, p = 2)
#> [[1]]
#> [1] 3 4 5
#>
#> [[2]]
#> [1] 2 4 5
#>
#> [[3]]
#> [1] 2 3 5
#>
#> [[4]]
#> [1] 2 3 4
#>
#> [[5]]
#> [1] 1 4 5
#>
#> [[6]]
#> [1] 1 3 5
#>
#> [[7]]
#> [1] 1 3 4
#>
#> [[8]]
#> [1] 1 2 5
#>
#> [[9]]
#> [1] 1 2 4
#>
#> [[10]]
#> [1] 1 2 3In order to use the ?resample function, we need to set
method = 'leave_p_out'.
#Leave-p-out sampling
obj = resample(
x = 5,
n = 2,
method = "leave_p_out"
)
#Print
print(obj)
#>
#> 10 samples taken from a population of 5 elements by using leave-p-out
#> sampling.
#>
#> sampleNumber sample sampleSize holdoutSize
#> 1 1 3, 4, 5 3 2
#> 2 2 2, 4, 5 3 2
#> 3 3 2, 3, 5 3 2
#> 4 4 2, 3, 4 3 2
#> 5 5 1, 4, 5 3 2
#> ...
#Samples
getSamples(obj)
#> [[1]]
#> [1] 3 4 5
#>
#> [[2]]
#> [1] 2 4 5
#>
#> [[3]]
#> [1] 2 3 5
#>
#> [[4]]
#> [1] 2 3 4
#>
#> [[5]]
#> [1] 1 4 5
#>
#> [[6]]
#> [1] 1 3 5
#>
#> [[7]]
#> [1] 1 3 4
#>
#> [[8]]
#> [1] 1 2 5
#>
#> [[9]]
#> [1] 1 2 4
#>
#> [[10]]
#> [1] 1 2 3
#Holdout Data
getHoldOutSamples(obj)
#> [[1]]
#> [1] 1 2
#>
#> [[2]]
#> [1] 1 3
#>
#> [[3]]
#> [1] 1 4
#>
#> [[4]]
#> [1] 1 5
#>
#> [[5]]
#> [1] 2 3
#>
#> [[6]]
#> [1] 2 4
#>
#> [[7]]
#> [1] 2 5
#>
#> [[8]]
#> [1] 3 4
#>
#> [[9]]
#> [1] 3 5
#>
#> [[10]]
#> [1] 4 5
#Plot
plot(obj)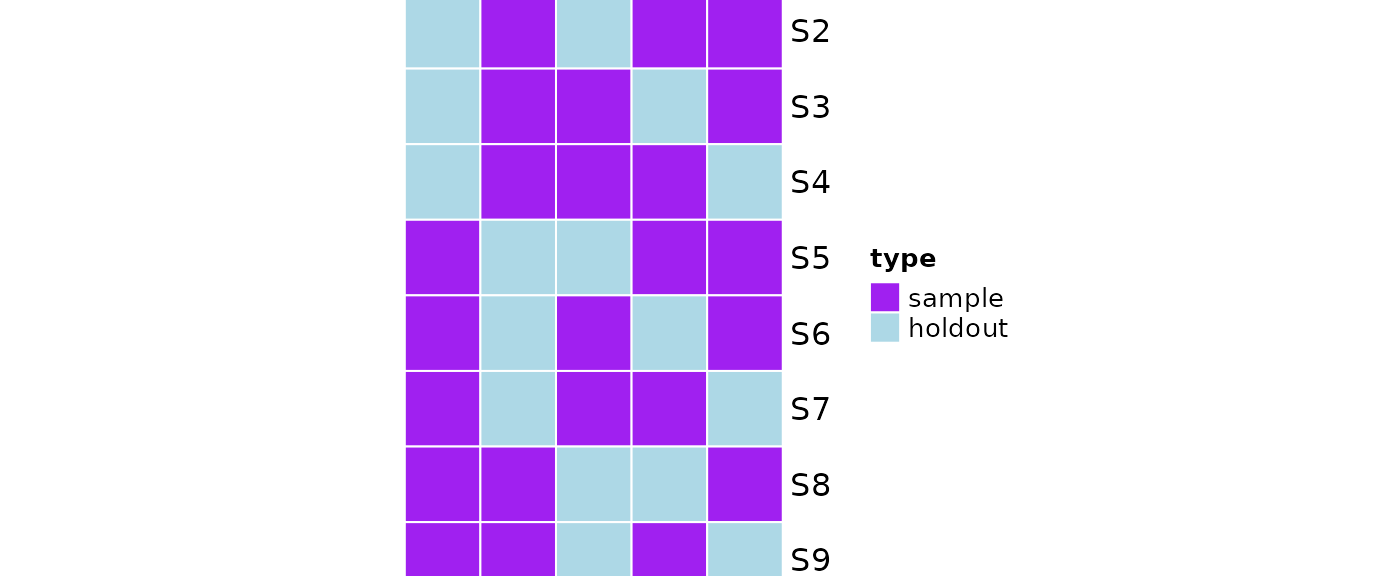
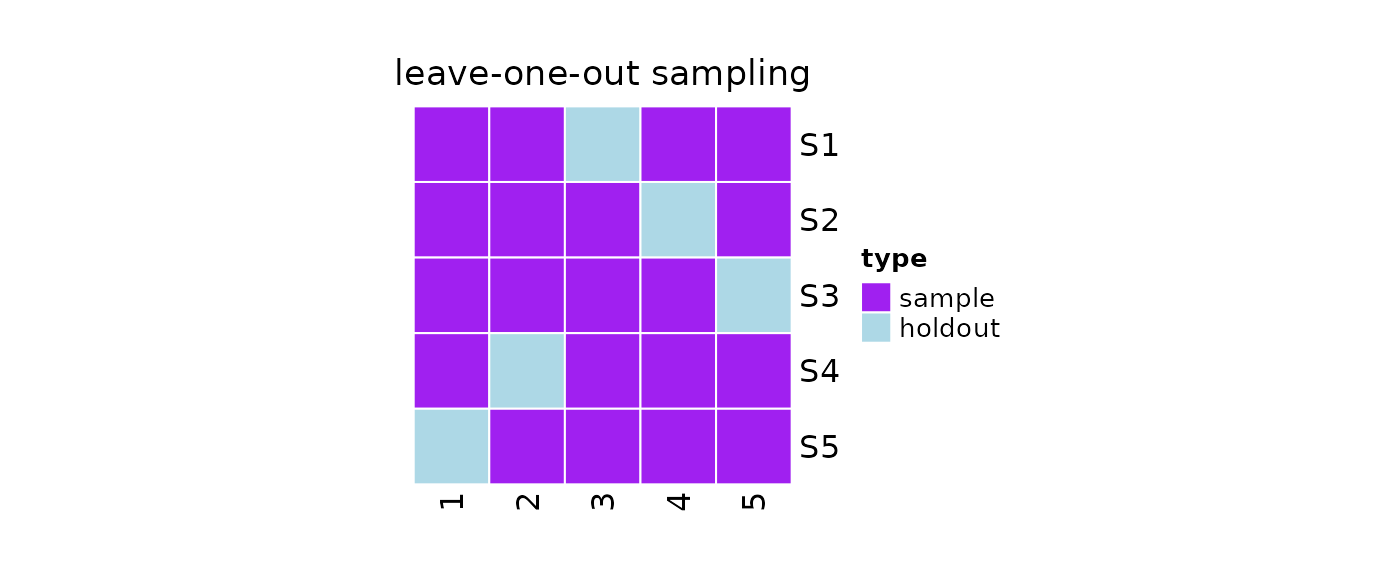
Leave-one-out Sampling
The leave-one-out is a particular case of the
leave-p-out, where p = 1. Similarly to
leave-p-out, leave-one-out resampling is an exhaustive
resampling technique, in which samples of size 1 are
repeatedly taken from the population until each element in the
population is considered as the holdout data.
?repeatedLeaveOneOut returns a list of length
N where each element is a sample obtained by removing the
holdout data:
#Take one sample leaving out 1 element
repeatedLeaveOneOut(N = 5)
#> [[1]]
#> [1] 1 3 4 5
#>
#> [[2]]
#> [1] 1 2 3 4
#>
#> [[3]]
#> [1] 2 3 4 5
#>
#> [[4]]
#> [1] 1 2 3 5
#>
#> [[5]]
#> [1] 1 2 4 5In order to use the ?resample function, we need to set
method = 'leave_one_out'.
#Leave-one-out sampling
obj = resample(
x = 5,
method = "leave_one_out"
)
#Print
print(obj)
#>
#> 5 samples taken from a population of 5 elements by using leave-one-out
#> sampling.
#>
#> sampleNumber sample sampleSize holdoutSize
#> 1 1 1, 2, 4, 5 4 1
#> 2 2 1, 2, 3, 5 4 1
#> 3 3 1, 2, 3, 4 4 1
#> 4 4 1, 3, 4, 5 4 1
#> 5 5 2, 3, 4, 5 4 1
#Samples
getSamples(obj)
#> [[1]]
#> [1] 1 2 4 5
#>
#> [[2]]
#> [1] 1 2 3 5
#>
#> [[3]]
#> [1] 1 2 3 4
#>
#> [[4]]
#> [1] 1 3 4 5
#>
#> [[5]]
#> [1] 2 3 4 5
#Holdout Data
getHoldOutSamples(obj)
#> [[1]]
#> [1] 3
#>
#> [[2]]
#> [1] 4
#>
#> [[3]]
#> [1] 5
#>
#> [[4]]
#> [1] 2
#>
#> [[5]]
#> [1] 1
#Plot
plot(obj)