Introduction
This article is used to benchmark the evaluations obtained by renoir for classification problems against external results.
Data
Different public datasets already used in classification problems are considered:
- The breast cancer data from the University of Wisconsin Hospitals
- The heart-disease data from the Cleveland Clinic Foundation
Data was retrieved from the UC Irvine Machine Learning Repository website.
Setup
Firstly, we load renoir and other needed packages:
library(renoir)
#> Warning: package 'Matrix' was built under R version 4.2.3
library(plotly)
library(htmltools)Learning Methods
Now we retrieve the ids of all the supported learners. A list of
supported methods is available through the
?list_supported_learning_methods function call.
#list methods
learning.methods = list_supported_learning_methods(x = "classification")
#print in table
knitr::kable(x = learning.methods)| id | method | default_hyperparameters |
|---|---|---|
| lasso | generalized linear model via L1 penalized maximum likelihood (lasso penalty) | lambda |
| ridge | generalized linear model via L2 penalized maximum likelihood (ridge penalty) | lambda |
| elasticnet | generalized linear model via L1/L2 penalized maximum likelihood (elasticnet penalty) | lambda, …. |
| relaxed_lasso | generalized linear model via L1 penalized maximum likelihood (relaxed lasso penalty) | lambda, …. |
| relaxed_ridge | generalized linear model via L2 penalized maximum likelihood (relaxed ridge penalty) | lambda, …. |
| relaxed_elasticnet | generalized linear model via L1/L2 penalized maximum likelihood (relaxed elasticnet penalty) | lambda, …. |
| randomForest | random forest | ntree |
| gbm | generalized boosted model | eta, ntree |
| linear_SVM | linear support vector machine | cost |
| polynomial_SVM | polynomial support vector machine | cost, ga…. |
| radial_SVM | radial support vector machine | cost, gamma |
| sigmoid_SVM | sigmoid support vector machine | cost, gamma |
| linear_NuSVM | linear nu-type support vector machine | nu |
| polynomial_NuSVM | polynomial nu-type support vector machine | nu, gamm…. |
| radial_NuSVM | radial nu-type support vector machine | nu, gamma |
| sigmoid_NuSVM | sigmoid nu-type support vector machine | nu, gamma |
| gknn | generalized k-nearest neighbours model | k |
| nsc | nearest shrunken centroid model | threshold |
We can extract the ids by selecting the id column.
#learning method ids
learning.methods.ids = learning.methods$idFrom the table, we can also see the default hyperparameters of the
methods. We create here our default values, and define a function to
dispatch them depending on the id in input:
#get hyperparameters
get_hp = function(id, y){
#Generalised Linear Models with Penalisation
lambda = 10^seq(3, -2, length=100)
# alpha = seq(0.1, 0.9, length = 9)
alpha = seq(0.1, 0.9, length = 5)
gamma = c(0, 0.25, 0.5, 0.75, 1)
#Random Forest
ntree = c(10, 50, 100, 250, 500)
#Generalised Boosted Regression Modelling
eta = c(0.3, 0.1, 0.01, 0.001)
#Support Vector Machines
cost = 2^seq(from = -5, to = 15, length.out = 5)
svm.gamma = 2^seq(from = -15, to = 3, length.out = 4)
degree = seq(from = 1, to = 3, length.out = 3)
#Note that for classification nu must be
#nu * length(y)/2 <= min(table(y)). So instead of
#fixing it as
# nu = seq(from = 0.1, to = 0.6, length.out = 5)
#we do
nu.to = floor((min(table(y)) * 2/length(y)) * 10) / 10
nu = seq(from = 0.1, to = nu.to, length.out = 5)
#kNN
k = seq(from = 1, to = 9, length.out = 5)
#Nearest Shrunken Centroid
threshold = seq(0, 2, length.out = 30)
#hyperparameters
out = switch(
id,
'lasso' = list(lambda = lambda),
'ridge' = list(lambda = lambda),
'elasticnet' = list(lambda = lambda, alpha = alpha),
'relaxed_lasso' = list(lambda = lambda, gamma = gamma),
'relaxed_ridge' = list(lambda = lambda, gamma = gamma),
'relaxed_elasticnet' = list(lambda = lambda, gamma = gamma, alpha = alpha),
'randomForest' = list(ntree = ntree),
'gbm' = list(eta = eta, ntree = ntree),
'linear_SVM' = list(cost = cost),
'polynomial_SVM' = list(cost = cost, gamma = svm.gamma, degree = degree),
'radial_SVM' = list(cost = cost, gamma = svm.gamma),
'sigmoid_SVM' = list(cost = cost, gamma = svm.gamma),
'linear_NuSVM' = list(nu = nu),
'polynomial_NuSVM' = list(nu = nu, gamma = svm.gamma, degree = degree),
'radial_NuSVM' = list(nu = nu, gamma = svm.gamma),
'sigmoid_NuSVM' = list(nu = nu, gamma = svm.gamma),
'gknn' = list(k = k),
'nsc' = list(threshold = threshold)
)
return(out)
}Performance metrics
A list of supported scoring metrics is available through the
?list_supported_performance_metrics function call.
#list metrics
performance.metrics = list_supported_performance_metrics(resp.type = "binomial")
#print in table
knitr::kable(x = performance.metrics)| id | name | problem |
|---|---|---|
| class | Misclassification Error | classification |
| acc | Accuracy | classification |
| auc | AUC | classification |
| f1s | F1 Score | classification |
| fbeta | F-beta Score | classification |
| precision | Precision | classification |
| sensitivity | Sensitivity | classification |
| jaccard | Jaccard Index | classification |
For this benchmark we want to select accuracy and precision, as these are the reported measure on the website.
#metric for tuning
performance.metric.id.tuning = "acc"
#metrics for evaluation
performance.metric.ids.evaluation = c("acc", "precision")Sampling Methods
A list of supported sampling methods is available through the
?list_supported_sampling_methods function call.
#list methods
sampling.methods = list_supported_sampling_methods()
#print in table
knitr::kable(x = sampling.methods)| id | name | supported |
|---|---|---|
| random | random sampling without replacement | stratification, balance |
| bootstrap | random sampling with replacement | stratification, balance |
| cv | cross-validation | stratification |
We decided to select the common scenario of a stratified 10-fold cross-validation for the tuning of hyperparameters, and repeated random sampling for the evaluation of the methodology.
#sampling for tuning
sampling.method.id.tuning = "cv"
#sampling for evaluation
sampling.method.id.evaluation = "random"Benchmark
We can now run our analyses.
Breast Cancer Data
The Breast Cancer database from the University of Wisconsin Hospitals was retrieved from the UC Irvine Machine Learning Repository website.
Load data
We load the data.
#load data
load(file = file.path("..", "..", "data-raw", "benchmark", "classification", "bcwo", "data", "bcwo_data.rda"))We need to set the response type.
#set response type
resp.type = "binomial"Setup
Learner
We can now create the related ?Learner objects.
#container
learners = list()
#loop
for(learning.method.id in learning.methods.ids){
#manual setup
learners[[learning.method.id]] = Learner(
tuner = tuner,
trainer = Trainer(id = learning.method.id),
forecaster = Forecaster(id = learning.method.id),
scorer = ScorerList(Scorer(id = performance.metric.id.tuning)),
selector = Selector(id = learning.method.id),
recorder = Recorder(id = learning.method.id),
marker = Marker(id = learning.method.id),
logger = Logger(level = "ALL")
)
}Evaluator
Finally, we need to set up the ?Evaluator.
#Evaluator
evaluator = Evaluator(
#Sampling strategy: stratified random sampling without replacement
sampler = Sampler(
method = "random",
k = 10L,
strata = y,
N = as.integer(length(y))
),
#Performance metric
scorer = ScorerList(
Scorer(id = performance.metric.ids.evaluation[1]),
Scorer(id = performance.metric.ids.evaluation[2])
)
)Analysis
Let’s create a directory to store the results.
#define path
outdir = file.path("..", "..", "data-raw", "benchmark", "classification", "bcwo", "analysis")
#create if not existing
if(!dir.exists(outdir)){dir.create(path = outdir, showWarnings = F, recursive = T)}Before running the analysis, we want to set a seed for the random
number generation (RNG). In fact, different R sessions have different
seeds created from current time and process ID by default, and
consequently different simulation results. By fixing a seed we ensure we
will be able to reproduce the results. We can specify a seed by calling
?set.seed.
In the code below, we set a seed before running the analysis for each considered learning method.
#container list
resl = list()
#loop
for(learning.method.id in learning.methods.ids){
#Each analysis can take hours, so we save data
#for future faster load
#path to file
fp.obj = file.path(outdir, paste0(learning.method.id,".rds"))
fp.sum = file.path(outdir, paste0("st_",learning.method.id,".rds"))
#check if exists
if(file.exists(fp.sum)){
#load
cat(paste0("Reading ", learning.method.id, "..."), sep = "")
resl[[learning.method.id]] = readRDS(file = fp.sum)
cat("DONE", sep = "\n")
} else {
cat(paste("Learning method:", learning.method.id), sep = "\n")
#Set a seed for RNG
set.seed(
#A seed
seed = 5381L, #a randomly chosen integer value
#The kind of RNG to use
kind = "Mersenne-Twister", #we make explicit the current R default value
#The kind of Normal generation
normal.kind = "Inversion" #we make explicit the current R default value
)
resl[[learning.method.id]] = renoir(
# filter,
#Training set size
npoints = 5,
# ngrid,
nmin = round(nrow(x)/2),
#Loop
looper = Looper(),
#Store
filename = "renoir",
outdir = NULL,
restore = TRUE,
#Learn
learner = learners[[learning.method.id]],
#Evaluate
evaluator = evaluator,
#Log
logger = Logger(level = "ALL", verbose = T),
#Data for training
hyperparameters = get_hp(id = learning.method.id, y = y),
x = x,
y = y,
weights = NULL,
offset = NULL,
resp.type = resp.type,
#Free space
rm.call = FALSE,
rm.fit = FALSE,
#Group results
grouping = TRUE,
#No screening
screening = NULL,
#Remove call from trainer to reduce space
keep.call = F
)
#save obj
saveRDS(object = resl[[learning.method.id]], file = fp.obj)
#create summary table
resl[[learning.method.id]] = renoir:::summary_table.RenoirList(resl[[learning.method.id]], key = c("id", "config"))
#save summary table
saveRDS(object = resl[[learning.method.id]], file = fp.sum)
cat("\n\n", sep = "\n")
}
}
#create summary table
resl = do.call(what = rbind, args = c(resl, make.row.names = F, stringsAsFactors = F))Performance
Let’s now plot the performance metrics for the opt and
1se configurations, considering the train, test, and full
set of data.
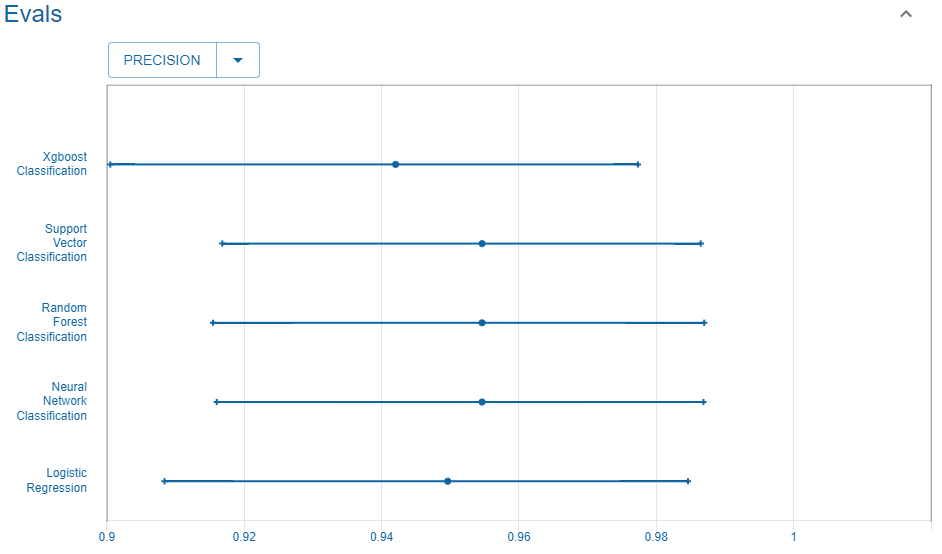
Precision
We consider the precision.
Reference
This is the precision reported in the UC Irvine Machine Learning Repository website that we use as reference.
Train
This is the precision score for the opt configuration
when considering the train set.
#plot
renoir:::plot.RenoirSummaryTable(
x = resl[resl$config == "opt",,drop=F],
measure = "precision",
set = "train",
interactive = T,
add.boxplot = F,
add.scores = F,
add.best = F,
key = c("id", "config")
)#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colors
#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colorsThis is the precision score for the 1se configuration
when considering the train set.
#plot
renoir:::plot.RenoirSummaryTable(
x = resl[resl$config == "1se",,drop=F], #select 1se config
measure = "precision",
set = "train",
interactive = T,
add.boxplot = F,
add.scores = F,
add.best = F,
key = c("id", "config")
)#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colors
#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colorsTest
This is the precision score for the opt configuration
when considering the test set.
#plot
renoir:::plot.RenoirSummaryTable(
x = resl[resl$config == "opt",,drop=F], #select opt config
measure = "precision",
set = "test",
interactive = T,
add.boxplot = F,
add.scores = F,
add.best = F,
key = c("id", "config")
)#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colors
#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colorsThis is the precision score for the 1se configuration
when considering the test set.
#plot
renoir:::plot.RenoirSummaryTable(
x = resl[resl$config == "1se",,drop=F], #select 1se config
measure = "precision",
set = "test",
interactive = T,
add.boxplot = F,
add.scores = F,
add.best = F,
key = c("id", "config")
)#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colors
#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colorsFull
This is the precision score for the opt configuration
when considering the full set.
#plot
renoir:::plot.RenoirSummaryTable(
x = resl[resl$config == "opt",,drop=F], #select opt config
measure = "precision",
set = "full",
interactive = T,
add.boxplot = F,
add.scores = F,
add.best = F,
key = c("id", "config")
)#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colors
#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colorsThis is the precision score for the 1se configuration
when considering the full set.
#plot
renoir:::plot.RenoirSummaryTable(
x = resl[resl$config == "1se",,drop=F], #select 1se config
measure = "precision",
set = "full",
interactive = T,
add.boxplot = F,
add.scores = F,
add.best = F,
key = c("id", "config")
)#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colors
#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
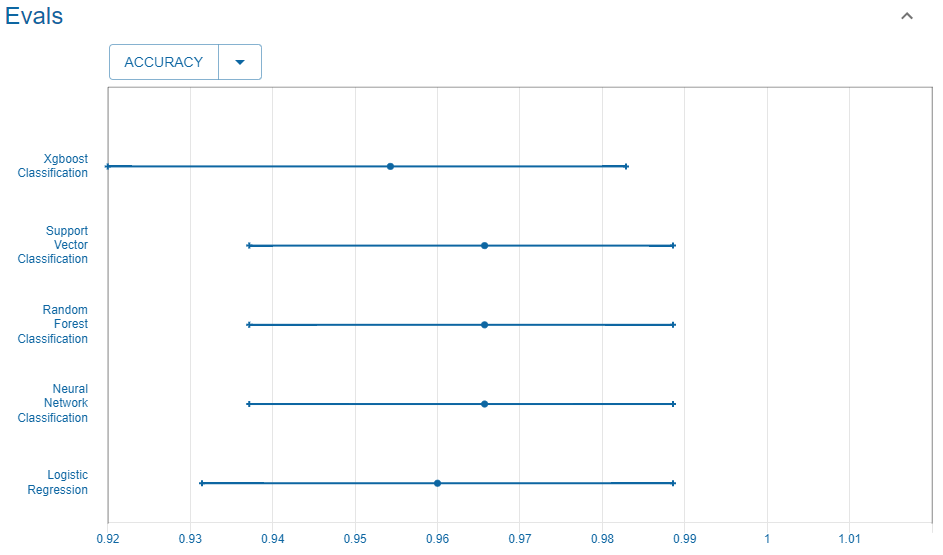
#> Returning the palette you asked for with that many colorsAccuracy
We consider here the accuracy score.
Reference
This is the accuracy reported in the UC Irvine Machine Learning Repository website that we use as reference.
Train
This is the accuracy score for the opt configuration
when considering the train set.
#plot
renoir:::plot.RenoirSummaryTable(
x = resl[resl$config == "opt",,drop=F], #select opt config
measure = "acc",
set = "train",
interactive = T,
add.boxplot = F,
add.scores = F,
add.best = F,
key = c("id", "config")
)#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colors
#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colorsThis is the accuracy score for the 1se configuration
when considering the train set.
#plot
renoir:::plot.RenoirSummaryTable(
x = resl[resl$config == "1se",,drop=F], #select 1se config
measure = "acc",
set = "train",
interactive = T,
add.boxplot = F,
add.scores = F,
add.best = F,
key = c("id", "config")
)#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colors
#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colorsTest
This is the accuracy score for the opt configuration
when considering the test set.
#plot
renoir:::plot.RenoirSummaryTable(
x = resl[resl$config == "opt",,drop=F], #select opt config
measure = "acc",
set = "test",
interactive = T,
add.boxplot = F,
add.scores = F,
add.best = F,
key = c("id", "config")
)#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colors
#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colorsThis is the accuracy score for the 1se configuration
when considering the test set.
#plot
renoir:::plot.RenoirSummaryTable(
x = resl[resl$config == "1se",,drop=F], #select 1se config
measure = "acc",
set = "test",
interactive = T,
add.boxplot = F,
add.scores = F,
add.best = F,
key = c("id", "config")
)#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colors
#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colorsFull
This is the accuracy score for the opt configuration
when considering the full set.
#plot
renoir:::plot.RenoirSummaryTable(
x = resl[resl$config == "opt",,drop=F], #select opt config
measure = "acc",
set = "full",
interactive = T,
add.boxplot = F,
add.scores = F,
add.best = F,
key = c("id", "config")
)#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colors
#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colorsThis is the accuracy score for the 1se configuration
when considering the full set.
#plot
renoir:::plot.RenoirSummaryTable(
x = resl[resl$config == "1se",,drop=F], #select 1se config
measure = "acc",
set = "full",
interactive = T,
add.boxplot = F,
add.scores = F,
add.best = F,
key = c("id", "config")
)#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colors
#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colorsHeart-Disease Data
The heart-disease database from the Cleveland Clinic Foundation was retrieved from the UC Irvine Machine Learning Repository website.
Setup
Learner
We can now create the related ?Learner objects.
#container
learners = list()
#loop
for(learning.method.id in learning.methods.ids){
#manual setup
learners[[learning.method.id]] = Learner(
tuner = tuner,
trainer = Trainer(id = learning.method.id),
forecaster = Forecaster(id = learning.method.id),
scorer = ScorerList(Scorer(id = performance.metric.id.tuning)),
selector = Selector(id = learning.method.id),
recorder = Recorder(id = learning.method.id, logger = Logger(level = "ALL", verbose = T)),
marker = Marker(id = learning.method.id, logger = Logger(level = "ALL", verbose = T)),
logger = Logger(level = "ALL")
)
}Evaluator
Finally, we need to set up the ?Evaluator.
#Evaluator
evaluator = Evaluator(
#Sampling strategy: stratified random sampling without replacement
sampler = Sampler(
method = "random",
k = 10L,
strata = y,
N = as.integer(length(y))
),
#Performance metric
scorer = ScorerList(
Scorer(id = performance.metric.ids.evaluation[1]),
Scorer(id = performance.metric.ids.evaluation[2])
)
)Analysis
Let’s create a directory to store the results.
#define path
outdir = file.path("..", "..", "data-raw", "benchmark", "classification", "heart_disease", "analysis")
#create if not existing
if(!dir.exists(outdir)){dir.create(path = outdir, showWarnings = F, recursive = T)}Before running the analysis, we want to set a seed for the random
number generation (RNG). In fact, different R sessions have different
seeds created from current time and process ID by default, and
consequently different simulation results. By fixing a seed we ensure we
will be able to reproduce the results. We can specify a seed by calling
?set.seed.
In the code below, we set a seed before running the analysis for each considered learning method.
#container list
resl = list()
#loop
for(learning.method.id in learning.methods.ids){
#Each analysis can take hours, so we save data
#for future faster load
#path to file
fp.obj = file.path(outdir, paste0(learning.method.id,".rds"))#fit object
fp.sum = file.path(outdir, paste0("st_",learning.method.id,".rds"))#summary table
#check if exists
if(file.exists(fp.sum)){
#load
cat(paste0("Reading ", learning.method.id, "..."), sep = "")
resl[[learning.method.id]] = readRDS(file = fp.sum)
cat("DONE", sep = "\n")
} else {
cat(paste("Learning method:", learning.method.id), sep = "\n")
#Set a seed for RNG
set.seed(
#A seed
seed = 5381L, #a randomly chosen integer value
#The kind of RNG to use
kind = "Mersenne-Twister", #we make explicit the current R default value
#The kind of Normal generation
normal.kind = "Inversion" #we make explicit the current R default value
)
resl[[learning.method.id]] = renoir(
# filter,
#Training set size
npoints = 5,
# ngrid,
nmin = round(nrow(x)/2),
#Loop
looper = Looper(),
#Store
filename = "renoir",
outdir = NULL,
restore = TRUE,
#Learn
learner = learners[[learning.method.id]],
#Evaluate
evaluator = evaluator,
#Log
logger = Logger(level = "ALL", verbose = T),
#Data for training
hyperparameters = get_hp(id = learning.method.id, y = y),
x = x,
y = y,
weights = NULL,
offset = NULL,
resp.type = resp.type,
#space
rm.call = FALSE,
rm.fit = FALSE,
#Group results
grouping = TRUE,
#No screening
screening = NULL,
#Remove call from trainer to reduce space
keep.call = F
)
#save
saveRDS(object = resl[[learning.method.id]], file = fp.obj)
#create summary table
resl[[learning.method.id]] = renoir:::summary_table.RenoirList(resl[[learning.method.id]], key = c("id", "config"))
#save summary table
saveRDS(object = resl[[learning.method.id]], file = fp.sum)
cat("\n\n", sep= "\n")
}
}
#create summary table
resl = do.call(what = rbind, args = c(resl, make.row.names = F, stringsAsFactors = F))Performance
Let’s now plot the performance metrics for the opt and
1se configurations, considering the train, test, and full
set of data.
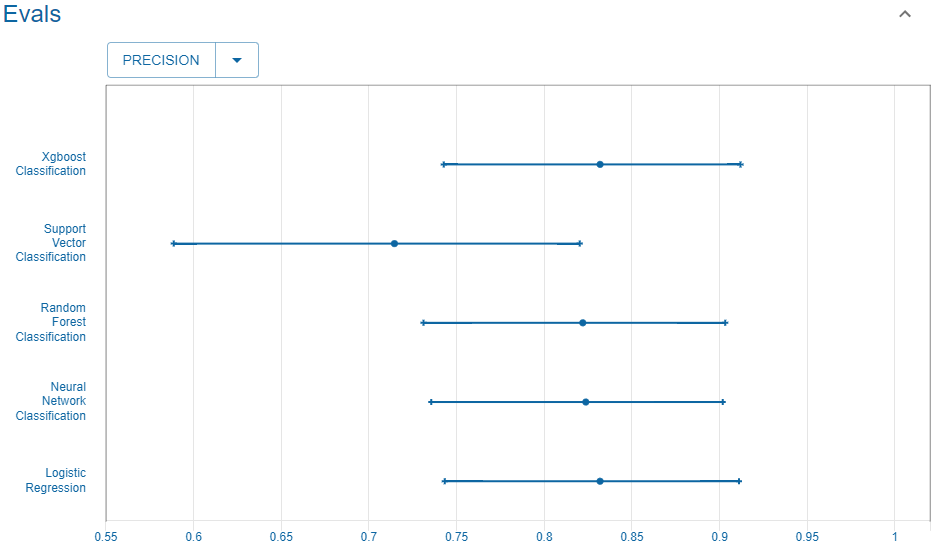
Precision
We consider the precision.
Reference
This is the precision reported in the UC Irvine Machine Learning Repository website that we use as reference.
Train
This is the precision score for the opt configuration
when considering the train set.
#plot
renoir:::plot.RenoirSummaryTable(
x = resl[resl$config == "opt",,drop=F], #select opt config
measure = "precision",
set = "train",
interactive = T,
add.boxplot = F,
add.scores = F,
add.best = F,
key = c("id", "config")
)#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colors
#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colorsThis is the precision score for the 1se configuration
when considering the train set.
#plot
renoir:::plot.RenoirSummaryTable(
x = resl[resl$config == "1se",,drop=F], #select 1se config
measure = "precision",
set = "train",
interactive = T,
add.boxplot = F,
add.scores = F,
add.best = F,
key = c("id", "config")
)#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colors
#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colorsTest
This is the precision score for the opt configuration
when considering the test set.
#plot
renoir:::plot.RenoirSummaryTable(
x = resl[resl$config == "opt",,drop=F], #select opt config
measure = "precision",
set = "test",
interactive = T,
add.boxplot = F,
add.scores = F,
add.best = F,
key = c("id", "config")
)#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colors
#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colorsThis is the precision score for the 1se configuration
when considering the test set.
#plot
renoir:::plot.RenoirSummaryTable(
x = resl[resl$config == "1se",,drop=F], #select 1se config
measure = "precision",
set = "test",
interactive = T,
add.boxplot = F,
add.scores = F,
add.best = F,
key = c("id", "config")
)#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colors
#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colorsFull
This is the precision score for the opt configuration
when considering the full set.
#plot
renoir:::plot.RenoirSummaryTable(
x = resl[resl$config == "opt",,drop=F], #select opt config
measure = "precision",
set = "full",
interactive = T,
add.boxplot = F,
add.scores = F,
add.best = F,
key = c("id", "config")
)#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colors
#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colorsThis is the precision score for the 1se configuration
when considering the full set.
#plot
renoir:::plot.RenoirSummaryTable(
x = resl[resl$config == "1se",,drop=F], #select 1se config
measure = "precision",
set = "full",
interactive = T,
add.boxplot = F,
add.scores = F,
add.best = F,
key = c("id", "config")
)#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colors
#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
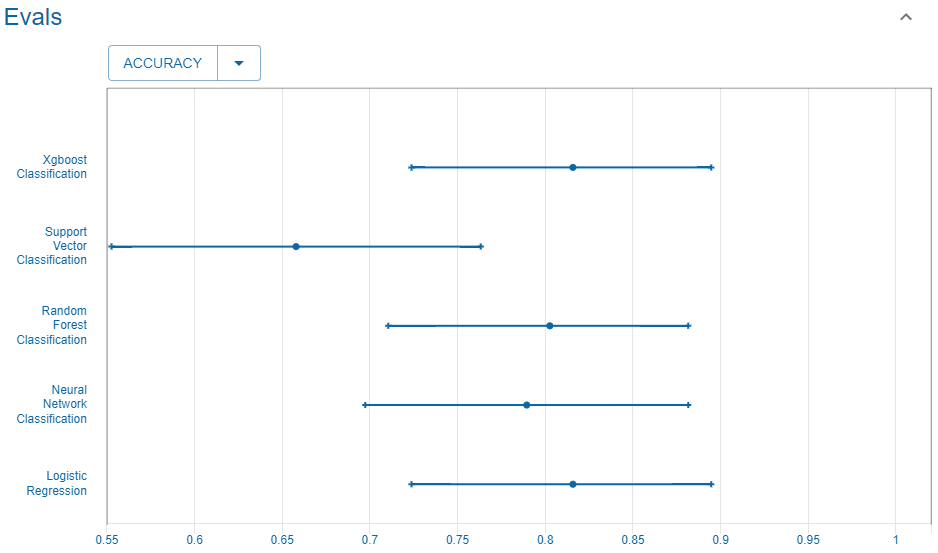
#> Returning the palette you asked for with that many colorsAccuracy
We consider here the accuracy score.
Reference
This is the accuracy reported in the UC Irvine Machine Learning Repository website that we use as reference.
Train
This is the accuracy score for the opt configuration
when considering the train set.
#plot
renoir:::plot.RenoirSummaryTable(
x = resl[resl$config == "opt",,drop=F], #select opt config
measure = "acc",
set = "train",
interactive = T,
add.boxplot = F,
add.scores = F,
add.best = F,
key = c("id", "config")
)#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colors
#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colorsThis is the accuracy score for the 1se configuration
when considering the train set.
#plot
renoir:::plot.RenoirSummaryTable(
x = resl[resl$config == "1se",,drop=F], #select 1se config
measure = "acc",
set = "train",
interactive = T,
add.boxplot = F,
add.scores = F,
add.best = F,
key = c("id", "config")
)#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colors
#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colorsTest
This is the accuracy score for the opt configuration
when considering the test set.
#plot
renoir:::plot.RenoirSummaryTable(
x = resl[resl$config == "opt",,drop=F], #select opt config
measure = "acc",
set = "test",
interactive = T,
add.boxplot = F,
add.scores = F,
add.best = F,
key = c("id", "config")
)#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colors
#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colorsThis is the accuracy score for the 1se configuration
when considering the test set.
#plot
renoir:::plot.RenoirSummaryTable(
x = resl[resl$config == "1se",,drop=F], #select 1se config
measure = "acc",
set = "test",
interactive = T,
add.boxplot = F,
add.scores = F,
add.best = F,
key = c("id", "config")
)#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colors
#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colorsFull
This is the accuracy score for the opt configuration
when considering the full set.
#plot
renoir:::plot.RenoirSummaryTable(
x = resl[resl$config == "opt",,drop=F], #select opt config
measure = "acc",
set = "full",
interactive = T,
add.boxplot = F,
add.scores = F,
add.best = F,
key = c("id", "config")
)#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colors
#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colorsThis is the accuracy score for the 1se configuration
when considering the full set.
#plot
renoir:::plot.RenoirSummaryTable(
x = resl[resl$config == "1se",,drop=F], #select 1se config
measure = "acc",
set = "full",
interactive = T,
add.boxplot = F,
add.scores = F,
add.best = F,
key = c("id", "config")
)#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colors
#> Warning in RColorBrewer::brewer.pal(N, "Set2"): n too large, allowed maximum for palette Set2 is 8
#> Returning the palette you asked for with that many colors